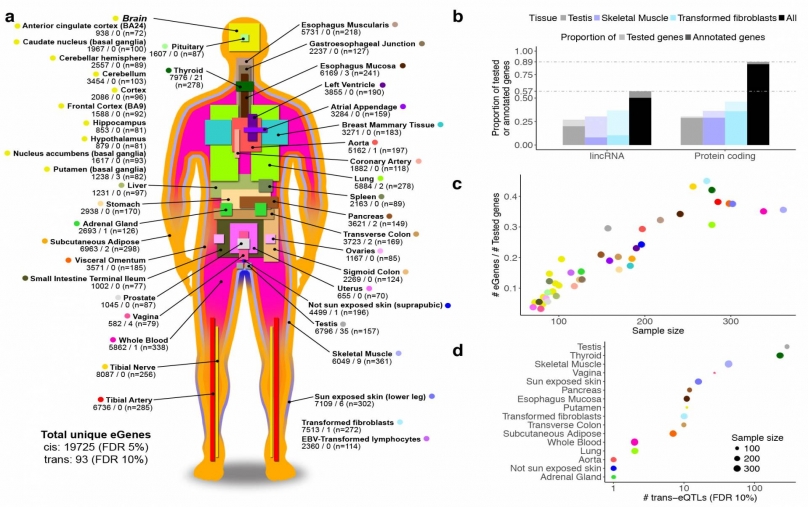
An international team of researchers reached a major milestone in decoding the human genome by linking genes across all chromosome of many individual people to specific tissues and disease processes. Using tissue samples donated from 449 people, the researchers linked nearly 20,000 genes to 44 tissue types. In the illustration, each tissue type is followed by the number of genes whose level of activity is controlled by nearby genes on the same chromosome (cis); those whose activity is associated with genes on other chromosomes (trans); and the number of tissue samples studied. Illustration courtesy of the GTEx Consortium
Engelhardt’s group was responsible for mapping associations between genetic variants and gene expression levels on different chromosomes, a connection known as “trans-expression quantitative trait loci (trans-eQTLS).” In contrast, cis-eQTLs — which account for the majority of genetic variation that affects gene expression — regulate genes located nearby on the same chromosome. Trans-eQTLs in particular have proven especially difficult to identify because of their biological and statistical complexity, Engelhardt said, but they might hold clues for explaining complex traits in a more comprehensive way than cis-eQTLs.
Engelhardt and her group’s role in the study included mapping and interpreting trans-eQTLs that they identified in the tissue samples. After clearing the samples of variance due to technical artifacts that could potentially confound the findings, they performed 3.5 trillion statistical tests against every mutation in the genome compared to every expressed gene in each of the 44 tissues. They used additional statistical techniques to correct for false positives in the data, which left them with several hundred trans-eQTLs. In the study, they additionally confirmed that nearby genetic variation in the form of cis-eQTLs affected expression of about 50 percent of genes in the samples. This work suggests, however, that this figure will climb to closer to 100 percent when more samples are added in the future.
“The extensive catalogue generated by the GTEx Consortium takes us one step closer to decoding the regulatory code of the genome,” said Yoav Gilad, a geneticist at the University of Chicago who was not involved in the study but was a scientific reviewer on the paper. “The consequences of genetic variation on gene expression are gradually becoming clearer.”
One trans-eQTL variant revealed in the study that was of particular interest was a mutation known to increase the risk of thyroid cancer. It is situated just next to a thyroid-specific transcription factor, a protein that regulates the rate of gene expression in the thyroid.
Prior to the study, the broad effects of the thyroid-specific transcription factor, called FOXE1, on transcription levels of genes were not well characterized. The researchers were able to replicate this finding by comparing the healthy thyroid tissues in GTEx to 500 samples taken from thyroid tumors, compiled by The Cancer Genome Atlas, and giving support to the extensive impact of FOXE1 on cellular state.
With these findings, “we can start to think about how to target specific genes for creating therapies for thyroid cancer,” Engelhardt said. “Many thyroid diseases will be impacted by changing the expression levels of the thyroid-specific transcription factor, so we want to investigate FOXE1 more carefully in future work.”
While the study represents a strong start for understanding how eQTLs affect gene regulation and expression, Engelhardt pointed out that she and her colleagues still do not have enough samples to understand trans-eQTLs as deeply as they would like. The GTEx Consortium is working on an analysis that includes almost three times as many samples as this current study. In addition, they hope soon to extend the project to new, underrepresented populations and build on existing efforts.
“The value of this dataset is in understanding and interpreting results in genome-wide studies,” Engelhardt said. “It’s already been extremely effective in understanding inherited diseases, and hopefully, as a resource, it continues to improve with more samples and better analyses.”